class: center, middle, inverse, title-slide .title[ # Stat 585 - Databases and R ] .author[ ### Heike Hofmann and Susan Vanderplas ] --- ## Outline - a bit about databases - linking R to a (local) data base - investigate the FARS data <br><br><br><br> Resources: - RStudio's [approach to databases](https://db.rstudio.com/) - `vignette("two-table", package="dplyr")` (for joining tables - material that was covered in Stat 579) --- ## Why databases? What is a database? - A collection of data - A set of rules to manipulate data <br><br> Why are databases important? - Efficient manipulation of large data sets - Convenient processing of data - Integration of multiple sources of data - Access to a shared resource --- ## Relational Databases - Database is collection of tables and links (normal form) - SQL (Structured Query Language) for querying - DBMS (Database Management System) and managing data --- ## Structured Query Language (SQL) - Structured Query Language (1970, E Codds) - Programming language used for accessing data in a database - ANSI standard since 1986, ISO standard since 1987 - Still some portability issues between software systems! - We'll mainly focus on SQL queries to access data --- ## Syntax - SQL is not case sensitive. - Some systems require a semi-colon `;` at the end of each line. The semi-colon can be used to separate each SQL statement in a system that allows multiple command to be executed in a call to the server. --- ## SQL SELECT statement https://dev.mysql.com/doc/refman/8.0/en/select.html ``` * SELECT [ALL | DISTINCT | DISTINCTROW ] [HIGH_PRIORITY] [STRAIGHT_JOIN] [SQL_SMALL_RESULT] [SQL_BIG_RESULT] [SQL_BUFFER_RESULT] [SQL_NO_CACHE] [SQL_CALC_FOUND_ROWS] * select_expr [, select_expr] ... [into_option] * [FROM table_references [PARTITION partition_list]] * [WHERE where_condition] * [GROUP BY {col_name | expr | position}, ... [WITH ROLLUP]] [HAVING where_condition] [WINDOW window_name AS (window_spec) [, window_name AS (window_spec)] ...] [ORDER BY {col_name | expr | position} [ASC | DESC], ... [WITH ROLLUP]] [LIMIT {[offset,] row_count | row_count OFFSET offset}] [into_option] [FOR {UPDATE | SHARE} [OF tbl_name [, tbl_name] ...] [NOWAIT | SKIP LOCKED] | LOCK IN SHARE MODE] [into_option] into_option: { INTO OUTFILE 'file_name' [CHARACTER SET charset_name] export_options | INTO DUMPFILE 'file_name' | INTO var_name [, var_name] ... } ``` --- ## Select .pull-left[ **Student** | ***ID*** | Name | Major | | ------------- |:-------------:| ------:| | 1234 | Never Ever | Math | | 4321 | Some Times | CS | ] .pull-right[ **Attendance** | ***ID*** | ***Date*** | Status | | ------------- |:-------------:| -------:| | 1234 | 02-Feb | absent | | 1234 | 09-Feb | absent | | 4321 | 02-Feb | present | | 4321 | 09-Feb | absent | ] <br> `Select * from student` leads to | *ID* | Name | Major | | ------------- |:-------------:| ------:| | 1234 | Never Ever | Math | | 4321 | Some Times | CS | --- ## Where .pull-left[ **Student** | ***ID*** | Name | Major | | ------------- |:-------------:| ------:| | 1234 | Never Ever | Math | | 4321 | Some Times | CS | ] .pull-right[ **Attendance** | ***ID*** | ***Date*** | Status | | ------------- |:-------------:| -------:| | 1234 | 02-Feb | absent | | 1234 | 09-Feb | absent | | 4321 | 02-Feb | present | | 4321 | 09-Feb | absent | ] <br> `*Select Name from student where Major='Math'` leads to | Name | | ------------- | | Never Ever | --- ## Aggregations .pull-left[ **Student** | ***ID*** | Name | Major | | ------------- |:-------------:| ------:| | 1234 | Never Ever | Math | | 4321 | Some Times | CS | ] .pull-right[ **Attendance** | ***ID*** | ***Date*** | Status | | ------------- |:-------------:| -------:| | 1234 | 02-Feb | absent | | 1234 | 09-Feb | absent | | 4321 | 02-Feb | present | | 4321 | 09-Feb | absent | ] <br> `Select ID, count(ID) from Attendance where Status='absent' group by ID` leads to | *ID* | Frequency | | ------------- |-------------:| | 1234 | 2 | | 4321 | 1 | --- ## Using SQL in R package `sqldf` provides a wrapper for sql statements in R: ``` sqldf::sqldf("Select ID, count(ID) from Attendance where Status='absent'") ``` Make sure to use a different set of quotes on the inside of a statement ```r sqldf::sqldf("select * from iris limit 5") ``` ``` ## Sepal.Length Sepal.Width Petal.Length Petal.Width Species ## 1 5.1 3.5 1.4 0.2 setosa ## 2 4.9 3.0 1.4 0.2 setosa ## 3 4.7 3.2 1.3 0.2 setosa ## 4 4.6 3.1 1.5 0.2 setosa ## 5 5.0 3.6 1.4 0.2 setosa ``` --- class: inverse # Your Turn - Use a SQL command wrapped in`sqldf` to create a table of Species (in the `iris` data) - Modify the command such that you only consider observations where petal length is less than 4 (Hint: sqldf does not like `.` in variable names - wrap the name in quotes) --- ## Additional Resources for SQL - W3Schools: http://www.w3schools.com/sql/default.asp - DBI package, RMySQL, RSQLite: http://cran.r-project.org/web/packages/RMySQL/RMySQL.pdf http://cran.r-project.org/web/packages/RSQLite/RSQLite.pdf --- ## Connecting to a database The package `DBI` allows to connect to various different DBMS, such as SQLite, MySQL, PostgreSQL, MonetDB, ... `dbConnect` and `dbDisconnect` `dbConnect` needs a DBMS specific driver, drivers have the form `SQLite(), MySQL(), ...` Connecting information depends on DBMS - e.g. MySQL needs a server, the name of the database, the user's information and generally a password --- ## SQLite - SQLite is a relational database management system - Unlike other DBMS SQLite is a server-less system with "zero-configuration" ```r download.file("https://github.com/Stat585-at-ISU/materials-2023/raw/main/data/fars2014", destfile = "fars2014", mode="wb") ``` connecting to the database: ```r library(dplyr) fars <- DBI::dbConnect(RSQLite::SQLite(), "fars2014") # connect to a database fars ``` ``` ## <SQLiteConnection> ## Path: /Users/hofmann/Documents/Teaching/Stat 585/Spring 2023/materials-2023/12_databases/fars2014 ## Extensions: TRUE ``` --- ## An example: FARS data - US Department of transportation is keeping a record of every accident that results in a fatality in the FARS Data base (fatal accident report system, http://www.nhtsa.gov/FARS) - FARS consists of 20+ tables consisting of various aspects of each accident Documentation at - http://www-nrd.nhtsa.dot.gov/Cats/listpublications.aspx?Id=J&ShowBy=DocType ```r DBI::dbListTables(fars) ``` ``` ## [1] "accidents" "person" "vehicle" ``` --- ## FARS 2014 - `fars2014` is a SQLite database of the 2014 data provided by FARS, consisting of three tables: `accidents`, `person`, and `vehicle` (make sure to download [`fars2014`](https://github.com/Stat585-at-ISU/materials-2023/raw/main/data/fars2014) from our github repo). ```r fars %>% tbl("accidents") ``` ``` ## # Source: table<accidents> [?? x 50] ## # Database: sqlite 3.40.0 [/Users/hofmann/Documents/Teaching/Stat 585/Spring 2023/materials-2023/12_databases/fars2014] ## STATE ST_CASE VE_TOTAL VE_FORMS PVH_INVL PEDS PERNOTMVIT ## <int> <int> <int> <int> <int> <int> <int> ## 1 1 10001 1 1 0 0 0 ## 2 1 10002 1 1 0 0 0 ## 3 1 10003 2 2 0 0 0 ## 4 1 10004 3 3 0 0 0 ## 5 1 10005 1 1 0 0 0 ## 6 1 10006 1 1 0 0 0 ## 7 1 10007 3 2 1 0 0 ## 8 1 10008 2 2 0 0 0 ## 9 1 10009 3 3 0 0 0 ## 10 1 10010 2 2 0 0 0 ## # ℹ more rows ## # ℹ 43 more variables: PERMVIT <int>, PERSONS <int>, ## # COUNTY <int>, CITY <int>, DAY <int>, MONTH <int>, ## # YEAR <int>, DAY_WEEK <int>, HOUR <int>, MINUTE <int>, ## # NHS <int>, ROAD_FNC <int>, ROUTE <int>, TWAY_ID <chr>, ## # TWAY_ID2 <chr>, MILEPT <int>, LATITUDE <dbl>, ## # LONGITUD <dbl>, SP_JUR <int>, HARM_EV <int>, … ``` --- ## Working with sql and dbplyr - `dbplyr` package works (almost) the same for local data frames as tables in a database - `dbplyr` functionality: `group_by`, `summarize`, `transform`, `filter`, `arrange`, `select` ```r fars %>% tbl("accidents") %>% group_by(DAY_WEEK, HOUR) %>% tally() ``` ``` ## # Source: SQL [?? x 3] ## # Database: sqlite 3.40.0 [/Users/hofmann/Documents/Teaching/Stat 585/Spring 2023/materials-2023/12_databases/fars2014] ## DAY_WEEK HOUR n ## <int> <int> <int> ## 1 1 0 296 ## 2 1 1 284 ## 3 1 2 322 ## 4 1 3 253 ## 5 1 4 175 ## 6 1 5 141 ## 7 1 6 124 ## 8 1 7 104 ## 9 1 8 94 ## 10 1 9 93 ## # ℹ more rows ``` --- ## Connecting to FARS Use `tbl` to connect to a specific table (dataset) in the database note: R does not load the data into the session unless forced with collect() ```r accidents <- tbl(fars, "accidents") accidents %>% dim() ``` ``` ## [1] NA 50 ``` ```r accidents %>% collect() %>% dim() ``` ``` ## [1] 29989 50 ``` ```r accidents ``` ``` ## # Source: table<accidents> [?? x 50] ## # Database: sqlite 3.40.0 [/Users/hofmann/Documents/Teaching/Stat 585/Spring 2023/materials-2023/12_databases/fars2014] ## STATE ST_CASE VE_TOTAL VE_FORMS PVH_INVL PEDS PERNOTMVIT ## <int> <int> <int> <int> <int> <int> <int> ## 1 1 10001 1 1 0 0 0 ## 2 1 10002 1 1 0 0 0 ## 3 1 10003 2 2 0 0 0 ## 4 1 10004 3 3 0 0 0 ## 5 1 10005 1 1 0 0 0 ## 6 1 10006 1 1 0 0 0 ## 7 1 10007 3 2 1 0 0 ## 8 1 10008 2 2 0 0 0 ## 9 1 10009 3 3 0 0 0 ## 10 1 10010 2 2 0 0 0 ## # ℹ more rows ## # ℹ 43 more variables: PERMVIT <int>, PERSONS <int>, ## # COUNTY <int>, CITY <int>, DAY <int>, MONTH <int>, ## # YEAR <int>, DAY_WEEK <int>, HOUR <int>, MINUTE <int>, ## # NHS <int>, ROAD_FNC <int>, ROUTE <int>, TWAY_ID <chr>, ## # TWAY_ID2 <chr>, MILEPT <int>, LATITUDE <dbl>, ## # LONGITUD <dbl>, SP_JUR <int>, HARM_EV <int>, … ``` --- ## Collecting results - `dbplyr` commands evaluate lazily, i.e. SQL commands are only generated and executed on demand - returns tibbles ```r accidents %>% filter(between(LONGITUD, -130, 0)) %>% ggplot(aes(LONGITUD, LATITUDE)) + geom_point(alpha = 0.5, size = 0.5) ``` --- 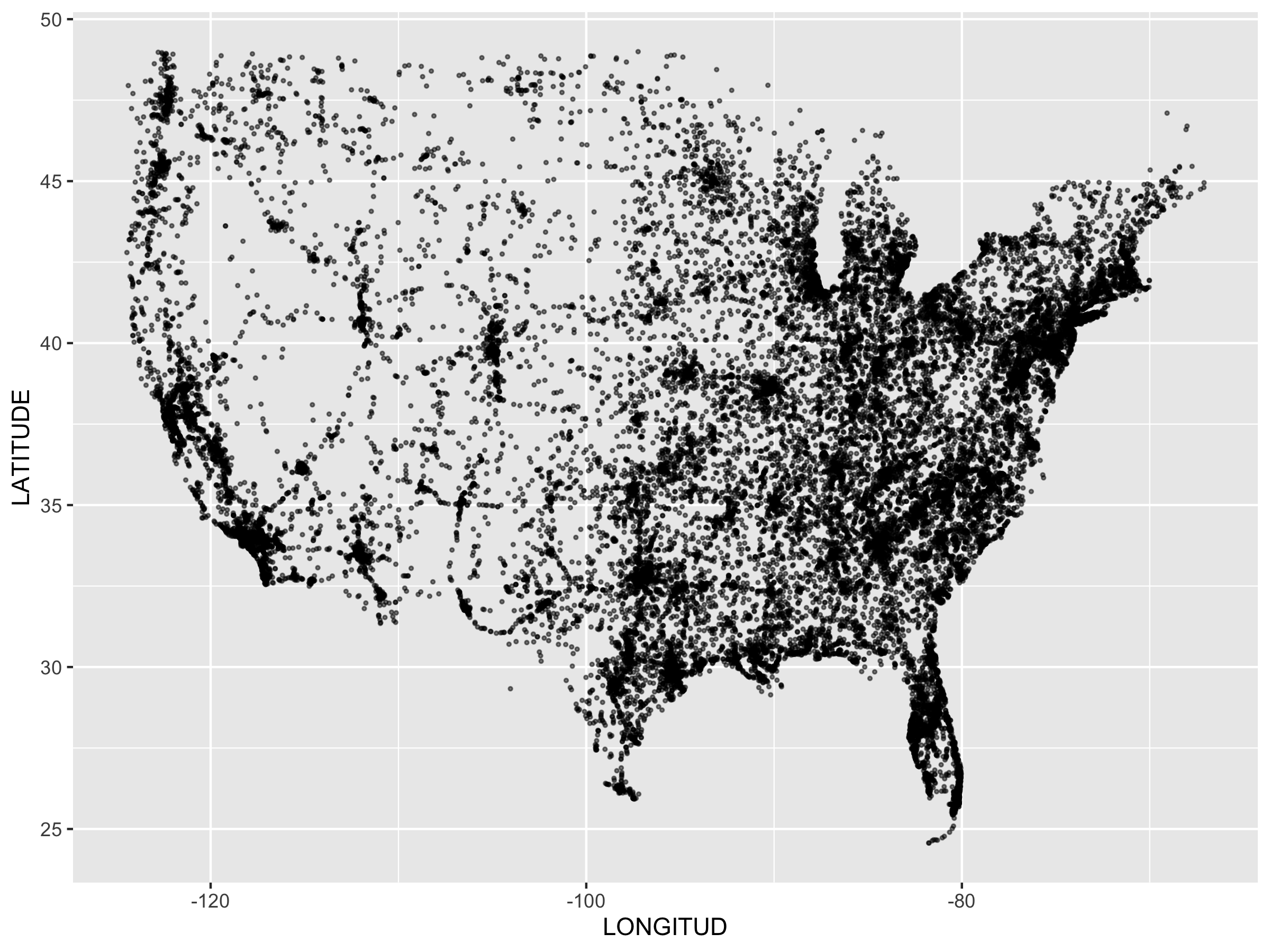<!-- --> --- class: inverse ## Your Turn (8 mins) Connect to the FARS database (using the SQLite database `fars2014`). Answer the following questions: - are there some days of the week where more accidents happen than on others (use variable `DAY_WEEK`)? - what time of the day do accidents happen (use variable `HOUR`)? - what is the number of accidents with at least one drunk driver (use variable `DRUNK_DR`)? --- ```r dayweek <- accidents %>% group_by(DAY_WEEK) %>% tally() dayweek %>% collect() %>% ggplot(aes(x = DAY_WEEK, weight=n)) + geom_bar() hours <- accidents %>% group_by(HOUR) %>% tally() hours %>% filter(HOUR < 25) %>% collect() %>% ggplot(aes(x = HOUR, weight=n)) + geom_bar() drunk <- accidents %>% group_by(DRUNK_DR) %>% tally() drunk %>% collect() %>% ggplot(aes(x = DRUNK_DR, weight=n)) + geom_bar() ``` --- ## Working with multiple tables - Only rarely single datasets are giving us all the answers we need - `left_join`, `right_join` work in the same way on databases as on local data frames. - think about dimensions ahead of time - the difference between a left and a right join is often a difference in multitude! --- class: inverse ## Your Turn (5 mins) Connect to the `person` table. Identify drivers (`PER_TYP` == 1, see [fars manual](https://www-fars.nhtsa.dot.gov/Help/Terms.aspx) ) and subset on them. - join the drivers and accidents tables. Which variable(s) do you use for the join? - Tally the number of accidents by day of the week (`DAY_WEEK`), hour of the day (`HOUR`) and gender (`SEX`). Visualize the results! --- ```r person <- tbl(fars, "person") drivers <- person %>% filter(PER_TYP == 1) driver_acc <- left_join(drivers, accidents) ``` ``` ## Joining with `by = join_by(STATE, ST_CASE, VE_FORMS, ## COUNTY, DAY, MONTH, HOUR, MINUTE, ROAD_FNC, HARM_EV, ## MAN_COLL, SCH_BUS)` ``` Note that there are a lot of variables with the same name - we actually include all of these variables --- ```r gg <- driver_acc %>% group_by(DAY_WEEK, HOUR, SEX) %>% tally() %>% filter(HOUR < 25, SEX < 8) %>% collect() %>% ggplot(aes(x = HOUR, y = n)) + geom_point(aes(colour = factor(SEX))) + facet_wrap(~DAY_WEEK, ncol=4) + scale_colour_brewer(palette="Set1") + theme(legend.position = "bottom") ``` --- ```r gg ``` 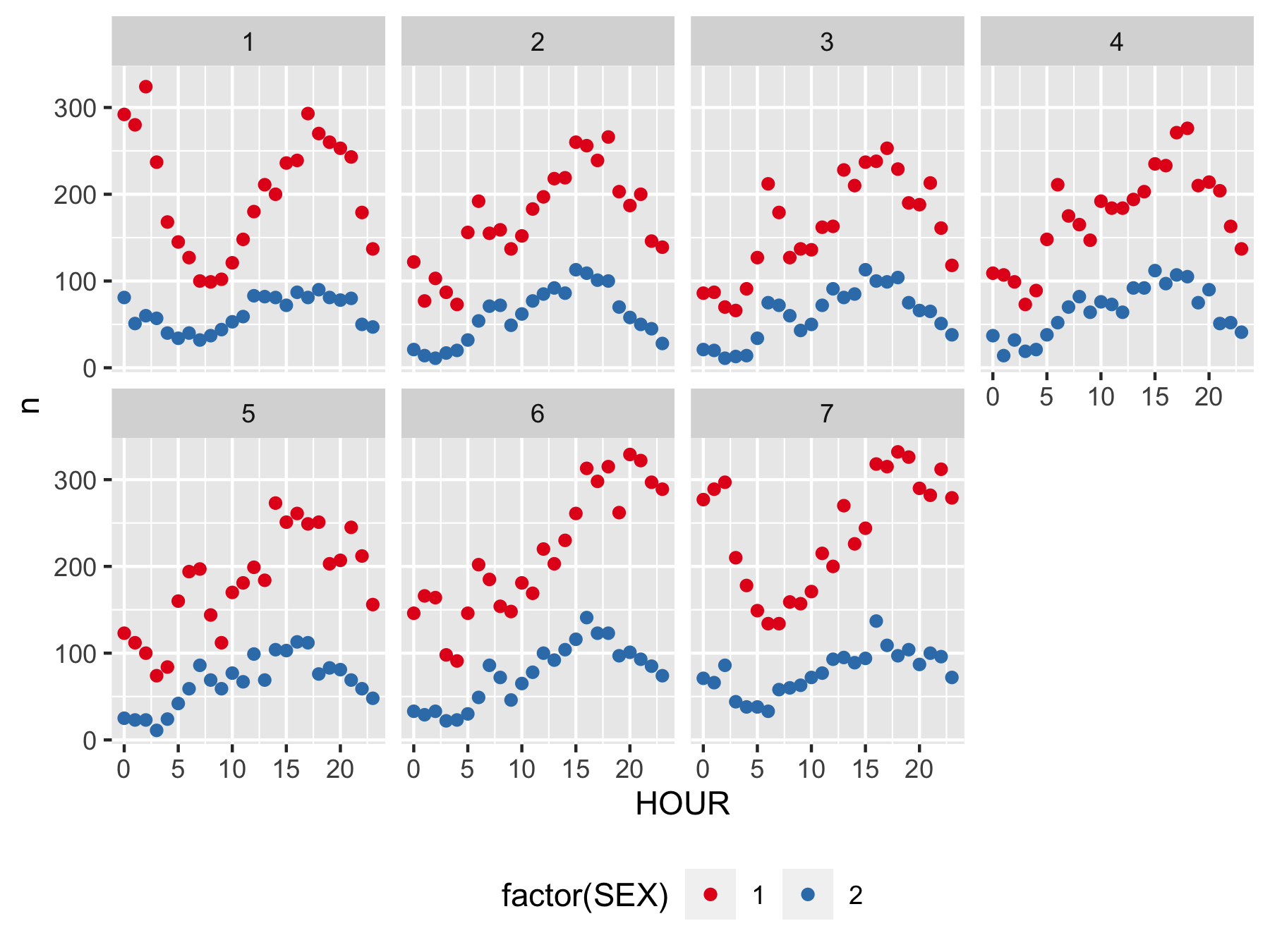<!-- --> --- class: inverse ## Your Turn **Creating a SQLite Database** The data for accidents in 2020 are published on the FARS website in csv form (in 20+ separate files). A subset of the tables is available from our github repo: [accident.csv, person.csv, and vehicle.csv]() Work through the RSQLite intro to create a new database `fars2020` from the three csv files: https://cran.r-project.org/web/packages/RSQLite/vignettes/RSQLite.html There should be an 'interesting' change in the pattern of accidents :( --- ```r library(DBI) fars2020 <- dbConnect(RSQLite::SQLite(), "fars2020.sqlite") accidents <- read.csv("../data/FARS-2020/accident.csv") dbWriteTable(fars2020, "accidents", accidents) person <- read.csv("../data/FARS-2020/person.csv") dbWriteTable(fars2020, "person", person) vehicle <- read.csv("../data/FARS-2020/vehicle.csv") dbWriteTable(fars2020, "vehicle", vehicle) dbListTables(fars2020) ``` ```r [1] "accidents" "person" "vehicle" ``` ```r fars2020 %>% tbl("accidents") %>% group_by(MONTH, DAY) %>% tally() %>% ggplot(aes(x = lubridate::dmy(paste(DAY, MONTH, "2023", sep="/")), y = n)) + geom_point() ```